Algorithm detective determines suitable neuro-model
In research, collecting data can be difficult, but fitting the data into a model that explains the observations is a challenge on another level. In neuroscience, many experiments measure the activity of neurons. However, to understand what exactly happens in a neuronal network, the data needs to be explained by a model. So, which model fits which data? A novel method developed by Pedro J. Gonçalves, Jan-Matthis Lueckmann, Michael Deistler, Jakob Macke and colleagues automates this process.
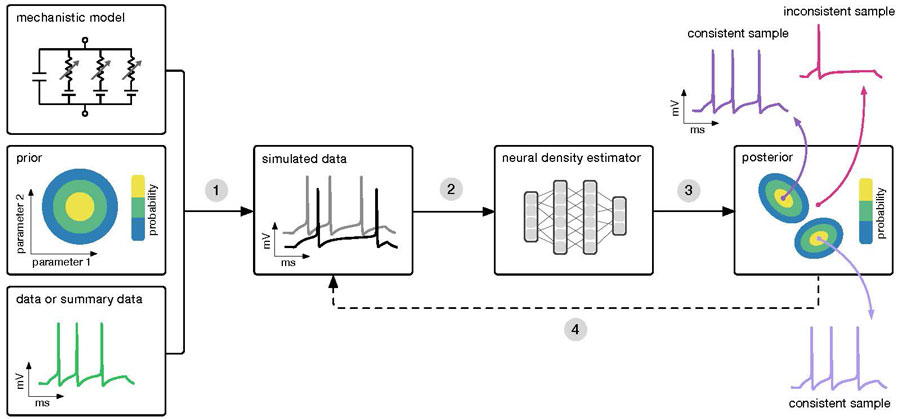
The algorithm (SNPE) takes three inputs: a model (i.e. computer code to simulate data from parameters), prior knowledge or constraints on parameters, and empirical data. SNPE runs simulations for a range of parameter values, and trains an artificial neural network to map any simulation result onto a range of possible parameters. After training on simulated data with known model parameters, SNPE provides the full space of parameters consistent with the empirical data and prior knowledge. By P. Goncalves, J. Lueckmann, M. Deistler and J. Macke
BN/ caesar, Scherrer et al./
The brain consists of a vast network formed by nerve cells. In order to understand processes in single neurons as well as interactions in networks of neurons, neural activity can be measured and a computational model can be used to understand the plethora of neuronal mechanisms. However, the identification of a suitable model, i.e. a model that matches the measured data, has so far often been very difficult, especially for complex, mechanistic models with many parameters.
Up until now, the possibilities of finding suitable models were limited: Either models were modified in painstaking work, until they could explain the measured data, or attempts were made to heuristically match models by using the “brute force” principle – i.e. the testing of all possible models. These approaches are either not systematic and prone to subjective criteria, or they are not efficient and are hence limited by the complexity of the model.
Pedro J. Gonçalves, Jan-Matthis Lueckmann, Michael Deistler, Jakob Macke and colleagues found a solution to this problem. They developed an algorithm that automatically determines suitable models based on the data observed in the experiment. Key to this approach is “Bayesian inference”: using the observed data and model simulations, the algorithm algorithmically identifies all suitable models that fit the measured data.
By using the novel algorithm, the scientific work is accelerated enormously. Remarkably, the new approach does not require any knowledge of the internal workings of the model and hence can be applied to models of various kinds and complexities. This makes it possible to use the algorithm in other areas of biomedical research as well.
Original publication
Gonçalves, Pedro J, Lueckmann, J., Deistler, M., Nonnenmacher,M., Öcal, K., Bassetto, G., Chintaluri, C., Podlaski, W.F., Haddad, S.A., Vogels,T.P., Greenberg, D.S. and Macke. J.H. “Training deep neural density estimators to identify mechanistic models of neural dynamics”. eLife. Oct. 27. 2020. Doi: 10.7554/eLife.56261




