Machine learning improves biological image analysis
International team of researchers develops algorithm that accelerates super-resolution microscopy
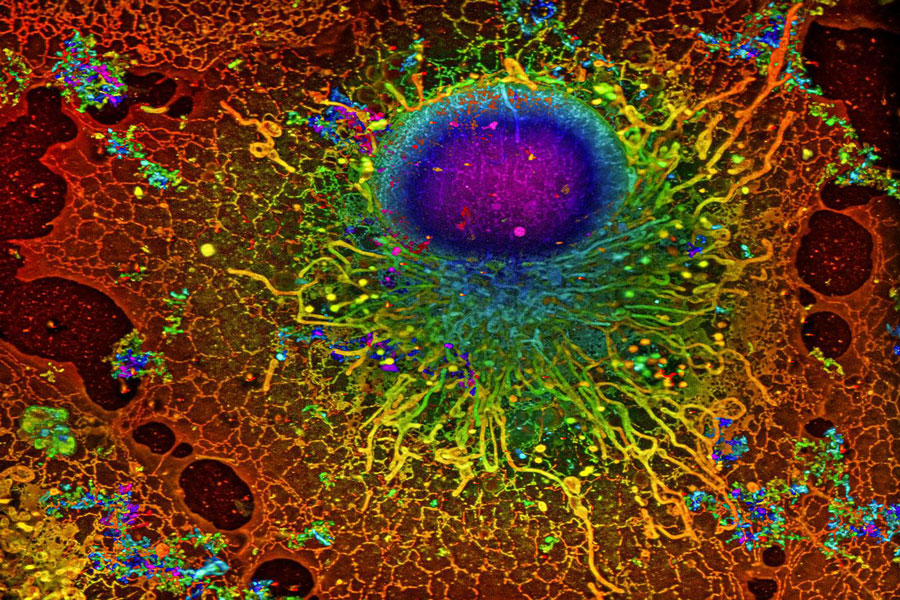
COS-7 cell imaged with SMLM; Image: Reconstruction by Artur Speiser et. al., data provided by Wesley Legant et. al.
Bernstein member involved: Jakob Macke
/Tübingen University/ Scientists use super-resolution microscopy to study previously undiscovered cellular worlds, revealing nanometer-scale details inside cells. This method revolutionized light microscopy and earned its inventors the 2014 Nobel Prize in Chemistry. In an international collaboration, AI researchers from Tübingen have now developed an algorithm that significantly accelerates this technology.
Single-molecule localization microscopy (SMLM) is a type of super-resolution microscopy. It involves labelling proteins of interest with fluorescent molecules and using light to activate only a few molecules at a time. Using this trick, multiple images of the same sample are acquired. To create a meaningful picture, a computer program unscrambles the data and compiles the complete image. While the technique can be used to locate molecules with high precision, it has one major drawback – it requires scientists to acquire a large number of images, which makes the process very time-consuming.
In an international collaboration, the team of Jakob Macke, Professor for Machine Learning in Science at the University of Tübingen, has developed a new algorithm that overcomes this limitation of SMLM. The joint work with the Ries Group at the European Molecular Biology Laboratory (EMBL) Heidelberg and Dr. Srinivas Turaga’s team at Janelia Research Campus (Virginia, USA) was published in Nature Methods.
Deep learning enables highly accurate single-molecule localisation
The DECODE (DEep COntext DEpendent) algorithm is based on deep learning: It uses a neural network that learns from training data. Instead of using real images, however, the network in this case is trained with synthetic data generated by a numerical simulation. By incorporating information about the microscopic setup and the imaging physics, the researchers achieved simulations that closely matched real-world acquisitions. “The neural network that we trained using simulated data can thus also detect and localize fluorophores in real images”, explains Artur Speiser, who, together with Lucas-Raphael Müller, was the lead author of the paper.
One of the benefits of DECODE is that it accurately detects and localizes fluorophores at higher densities than were previously possible. This means that fewer images are needed per sample. As a result, imaging speeds can be increased up to tenfold with minimal loss of resolution. In addition, DECODE can quantify uncertainties – so the network itself can detect when it is unsure of its localization.
Interdisciplinarity expands the perspectives of research
“This work is indicative of the approach of our Cluster of Excellence ‘Machine Learning: New Perspectives for Science’”, Macke says, whose chair is part of the Tübingen cluster. “We originally developed the ideas underlying the machine learning approach in a very different context, but through collaborating with experts in computational microscopy we were able to turn them into powerful methods for analyzing SMLM data.”
The team has also built a software package which implements the DECODE algorithm. “The software is simple to install and free to use, so we hope it will be useful for many scientists in the future”, adds Dr. Jonas Ries from EMBL.
Original publication
Artur Speiser, Lucas-Raphael Müller, Philipp Hoess, Ulf Matti, Christopher J. Obara, Wesley R. Legant, Anna Kreshuk, Jakob H. Macke, Jonas Ries & Srinivas C. Turaga: Deep learning enables fast and dense single-molecule localization with high accuracy. Nature Methods. https://doi.org/10.1038/s41592-021-01236-x (2021).




